Introduction
Since starting at DWP Digital, I’ve spent quite a bit of time working with some colleagues developing our analysis environments. We need to be able to control and adapt our environments quite rapidly so that we can use the latest and greatest tools, but we also need these environments to be robust and reproducible and to be deployed in a variety of hosting contexts including laptops and our secure internal environment. Containerisation technology, specifically Docker, has emerged as a key tool in this and I wrote a non-technical ‘official’ blog for DWP Digital which gained quite a bit of interest. This post covers the technical aspects of how what we actually did this which will hopefully be helpful to others in a similar situation by walking through an example codebase on GitHub at chapmandu2/datasci_docker.
Three components of reproducibility
There are three important aspects to reproducible data science:
- code
- data
- environment
Unless we have systems and processes in place to control and monitor changes to these three aspects, we won’t be able to reproduce our analysis. Data is perhaps the easiest aspect to control, since in many cases the data is static and just won’t change, but even so it is worth recording data time stamps or creation dates. If the data is a live feed then things become a lot more complex. Code version control is a critical component of data analysis and git has emerged as the de-facto standard in data science.
Open source is great, but also a challenge for reproducibility
Control of environments is more difficult. The diversity and rapid evolution of data science tools is a huge benefit in contemporary data science, but the plethora of libraries, packages and versions can also cause problems. A slight change in functionality between versions of one library can result in a change in the output of an analysis, even though the same code is used. Although environment management options are available for R and Python themselves (packrat for R, pipenv and conda for Python), these can still be problematic to use, especially if components outside of R/Python are important, for example system libraries.
Enter Docker
Docker has emerged as a way to manage data science environments. Docker containers can be thought of as very lightweight virtual machines, with the advantage that they can be relatively small and also fast to instantiate. A Docker container is derived from a Docker image, and an image can be built from a recipe file called a Dockerfile. This means that the code to build the Docker image can be version controlled in the same way as the analysis code, but also that the Docker image can be archived and retrieved in future so that the exact same environment can be used to run an analysis. The rest of this blog post describes one possible workflow for using Docker for environment management, that also integrates Makefiles and git for a very high level of reproducibility.
Overview of the workflow
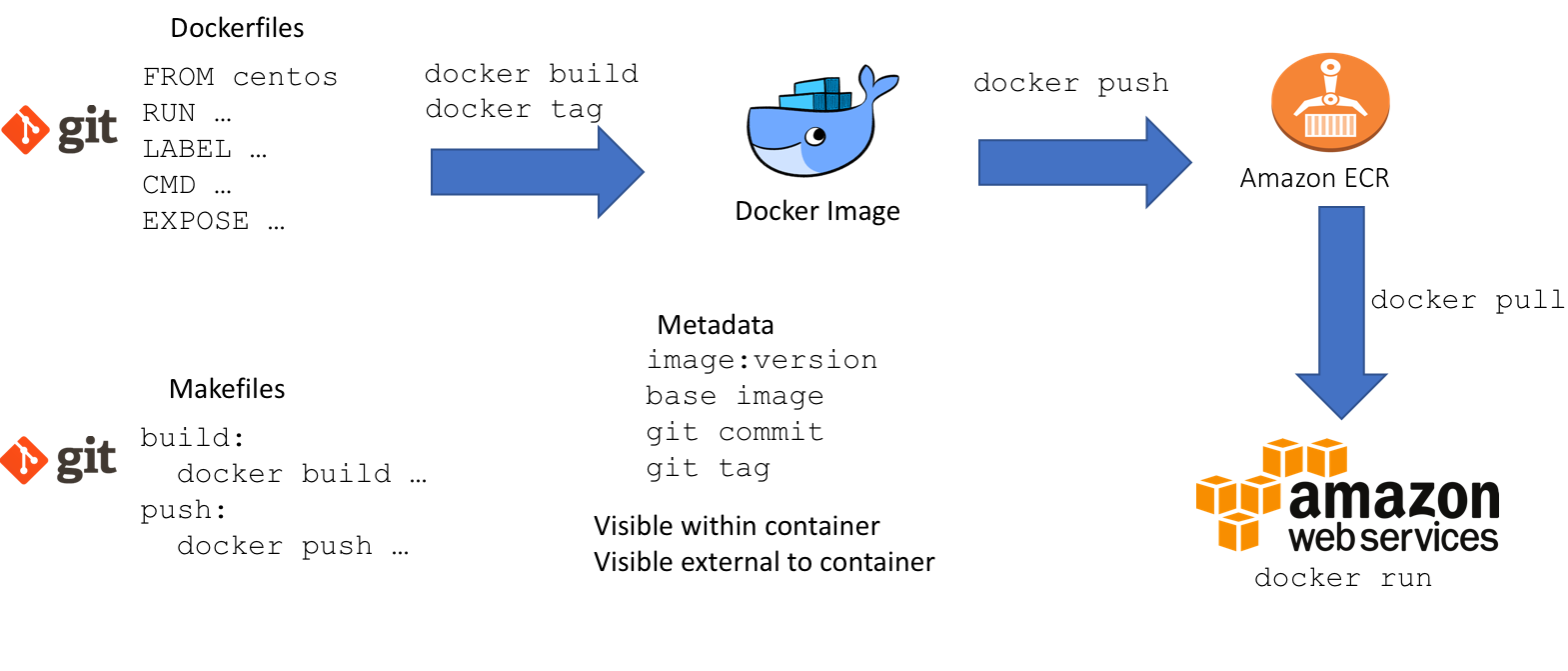
Images as layers
We built our images as layers which build on top of eachother:
- 00-docker: creates some foundations for capturing metadata
- 01-linux: installs the linux system libraries and applications such as R
- 02-r: installs a set of R packages which tend to be used across all projects
- 03-rstudio: installs RStudio Server
On top of these we then build further project specific images which share the components above, but add project specific requirements. For example, an NLP project might add various NLP packages. This gives a balance of having some consistency between projects, whilst allowing projects to have flexibility over the tools they have access to.
Dockerfiles as recipes
A Dockerfile is like a recipe for a cake. Just as a recipe species which ingredients in what order are required to make the cake mix, we can specify the exact instructions required to make a Docker image. Which directories have to be created, which software has to be installed, all of this is captured in a Dockerfile, and as this is just a text file it can be version controlled using git.
Makefiles as oven settings
If a Dockerfile is a recipe, then Makefiles are like the cooking instructions. To bake a cake we need to know how long we need to bake it for and at what temperature, otherwise the cake will turn out differently even though the cake mix was the same. The same Dockerfile will give a slightly different result depending on the parameters that are specified in the docker build and docker run commands. This is especially important for controlling how metadata is captured. Just like Dockerfiles, Makefiles can also be version controlled with git.
Container registries as a freezer
So we have our recipe and our cooking instructions, but if I bake a cake today it won’t be exactly the same as a cake baked in a year’s time: the eggs will be slightly different, I might be using a different oven. In the same way, even with our best efforts it is very difficult to guarantee that a docker image will be the same because the build process depends on various resources on the internet from which we download the software. Even if we try to specify exact software versions, it’s going to be difficult to guarantee an identical build. Therefore the only solution is to archive the Docker image itself, just as we might freeze a cake. To do this we can use a container registry such as Amazon’s Elastic Container Registry.
Metadata and git integration
We’ve gone to a lot of effort to ensure reproducibility, the last step is to capture metadata in our Docker image so that we know how it was created and what version it was. We want to link the Docker image back to the code that created it, and the way to do this is to link it to our version control system: git. There are two ways in which we need this metadata to be visible:
- from the outside: so we can choose which Docker image to use.
- from the inside: so that when we carry out our analysis we know which Docker image we’re using.
The way that this is managed in the example repository is as follows:
- the make command captures the current git commit and any git tags
- these are included in the
docker buildcommand as build arguments - the build arguments are captured in the Dockerfile as Docker labels that can be retrieved with a
docker inspectcommand (external metadata) - the build arguments are fed to a shell script (see 00-docker) which creates various audit files in
/etc/docker - the information in
/etc/dockeris then available to anything running within the Docker container (internal metadata)
In practice a message is shown when R is launched that shows the build information, and this message can also be included in output log files or within an RMarkdown document.
################################
# DOCKER BUILD INFORMATION
################################
This is docker image: ds-stage-02-r:v1.1
Built from git commit: 3d07371
See https://github.com/chapmandu2/datasci_docker/tree/3d07371 for Dockerfile code
See /etc/docker/docker_build_history.txt for provenance of this environment.
See /etc/docker for more information
################################
Workflow instructions
Clone the git repo
Assuming that you already have Docker installed on your computer, clone the git repository chapmandu2/datasci_docker from GitHub:
git clone https://github.com/chapmandu2/datasci_docker
Repo structure
Within the repo there is a Makefile and a readme and a number of directories each corresponding to an image. The Makefile at the top level is the master Makefile and contains the shared logic required to build all of the images. Each image also has its own Makefile which references the master Makefile. Feel free to look at the Makefile but you don’t need to understand the details at this stage.
Make the 00-docker image
Navigate to the ds-stage-00-docker directory and enter make build - this will build the first docker image which includes the metadata audit functionality. If you wish you can enter make run which will create and run a container from this image and you can look in the /etc/docker directory. The generate_docker_info.sh script is run during the docker build process to capture and store all of the metadata.
Make the 01-linux image
Next navigate to the ds-stage-01-linux directory and again enter make build to build the linux base image. This will take a minute or two depending on your internet connection as various software libraries are downloaded. Once finished, you can again enter make run to explore the image - check out the /etc/docker directory to see how the metadata has changed.
Make the 02-r image
Now navigate to the ds-stage-02-r directory do the same steps as above: make build to build the image and make run to run it. For a bit of fun try this command:
docker run -it --rm --entrypoint R ds-stage-02-r:latest
This will instantiate a container from the image as before, but rather than expose a bash prompt will run R and expose this. To the user it’s almost as quick as launching R running natively on their local system.
Make the 03-rstudio image
As before, navigate to the directory and enter make build and make run. This time, however, the container runs in detatched mode and exposes an RStudio Server instance which can be accessed from localhost:8787 in a browser and authenticated using rstudio:rstudio. Now we are running R within the RStudio Server IDE. It is this image which can be deployed onto a server, whether on the local network or on the cloud, allowing users to log in from their browser and use a reproducible data environment.
Make a project specific image
To make a project specific image, it is as simple as building on the contents of the ds-stage-11-project directory. Add any linux system requirements, such as libpng in this case, and any R packages to the relevant sections. You can be as sophisticated as you like here, but the best approach is to work out your installation process on a container of the base image first, and then transfer this process into the Dockerfile. I will try to add some more sophisticated examples later, we have images with Python also installed, difficult R installs such as the RStan, and R packages installed from github. Dockerhub is a good place to look for inspiration.
Docker registries
I have deliberately not included in this post the part of the workflow that involves pushing the Docker image to a registry. However, the code is present in the Makefile and it is just a case of specifying my-repo/my-image as the base image, and modifying the Amazon ECR section of the master Makefile (line 40). Then add a make push command to the sequence of make commands used thus far. Note that I set up some logic that required a git commit associated with the image to be tagged in order to be pushed to a repository.
The benefit of using a registry is that it makes it easier to deploy and share images, and that the build process can be carried out anywhere that the git repo and docker registry is accessible from. One useful option is to build images on an EC2 instance on AWS, since this has a very fast network connection that expidites the build process.
Potential improvements
The template shared here seems to have worked reasonably well but there is room for improvement. I am a complete beginner in using Make and whilst it’s very powerful, the Makefiles could be more cleanly constructed. In addition, shared parameters such as the registry address could be stored in a repo-wide yaml file rather than being buried within a Makefile. The main improvement, however, would be in how git tags and commit id’s are used. As it stands, changes to the Dockerfile could be made but not committed, and the resultant image would still have the current git commit associated with it. I left it like this for pragmatic reasons - when you are developing Docker containers it’s a pain to have to commit every time you attempt to build! So it’s important to remember this, and at the end when you think you’re done, to go back and build every image again with the same code.
Conclusions
In this blog post I have shared a workflow for developing reproducible data science environments using Docker. I hope that it useful to others in their research, and please let me know if you have any ideas for improvements!
Postscript
- There are RStudio and R images available on DockerHub under the rocker account. These can be more convenient starting points than a linux base image. In our case we needed Centos as the base Linux OS rather than Ubuntu, hence why we started from scratch.
- The example git repo was designed to be relatively quick to build for example purposes, it probably doesn’t include enough linux libraries or R packages to actually be useful. I’ll try to add a more useful example to the repo.